Equinox Graphics / Science Photo Library
In March 2013, China announced that two people had died from an infection of bird influenza. This particular strain of flu, known as H7N9, had not previously been seen in humans or animals, and there was no vaccine available. As the outbreak worsened, more people began to die. Scientists had faced a similar situation four years earlier when the H1N1 flu pandemic had emerged. Despite a record-breaking response by the pharmaceutical industry, creating a vaccine against H1N1 took too long to make a real impact. This time, against H7N9, some scientists were determined to act faster.
“We sprang into action,” says Sammy Farah, president of the Synthetic Genomics Vaccines unit at Synthetic Genomics in La Jolla, California, whose company had been preparing for this scenario since the 2009 outbreak.

Source: Synthetic Genomics
Bolyn Hubby (left) and Sammy Farah work at Synthetic Genomics, which produces flu vaccines using synthetic biology
Within five days, Synthetic Genomics, in partnership with Novartis, had made the ‘seed’ virus needed to initiate vaccine production — it usually takes four to six weeks or even longer to get to this point, says Farah. The vaccine soon went into phase I clinical trials and was stockpiled by the US government in December 2013, in time to fight a second wave of H7N9 infection if it were ever to reach US soil, which it did not.
By using synthetic biology, the company bypassed the old method of vaccine production using eggs. This was made possible by a forward-thinking move from China. When the Chinese public-health authorities announced that a deadly new strain of flu had surfaced, they simultaneously published the DNA sequence of the virus. This allowed Synthetic Genomics to download the crucial haemagglutinin (H) and neuraminidase (N) genes, synthesise them in the laboratory and combine them with the ‘backbone’ genes needed for viral growth and replication. The virus was then quickly grown in a cell-based vaccine production system developed by Novartis. Using the old technique, physical samples of the virus would need to be isolated and an attenuated version created and then grown in eggs before being shipped to manufacturers for vaccine production, also in eggs, a process that takes about five to six months.
Although the H7N9 outbreak did not reach pandemic proportions, Farah says that Synthetic Genomics and Novartis used the outbreak to demonstrate that they could now respond quickly in the event of a pandemic. “Once the sequence of the H7N9 genes was available, we could synthesise those genes within hours,” says Bolyn Hubby, head of research and development at Synthetic Genomics Vaccines, adding that this was a “simple and elegant application of synthetic biology”.
Once the sequence of the H7N9 genes was available, we could synthesise those genes within hours
Synthetic biology is being applied elsewhere too. From space travel to Mars, to drugs that can be manufactured on demand, researchers are turning to synthetic biology to solve the complex problems of the 21st century. The technology has been made possible by two advances: the recent ability to synthesise huge amounts of DNA extremely quickly, and the availability of immense computing power.
Life’s operating system
But when asked to define exactly what synthetic biology is, Farah and Hubby hesitate. “That’s a good question,” says Farah, chuckling. After some thought, he adds: “We think of it as the modular design of genomes, and therefore cells, for a particular function.” Synthetic biologists sometimes liken it to computer programming. “Often we use an operating-system analogy — DNA and cells are the operating system for life,” says Farah.
Synthetic biology is in some ways a sophisticated version of genetic engineering, which in its basic form has been around for decades. But there are differences. Old-fashioned genetic engineering adds or removes individual genes, which is like downloading or deleting a new piece of software on your computer. In contrast, synthetic biology rewrites your whole operating system and adds some novel code at the same time. “It is a matter of scale,” says Farah. In synthetic biology, whole new metabolic pathways, genes or even organisms are designed by using the principles of modern engineering. DNA sequences are written like code, synthesised in the lab and inserted into the cell. Traditional genetic-engineering techniques cannot deal with large gene constructs or whole genomes, says Farah.
One of the pioneers of synthetic biology was J. Craig Venter, who founded Synthetic Genomics. In 1995, Venter led the team that sequenced the first cellular genome, that of the bacteria Haemophilus influenzae
[1]
. And in 2001, he led a private project to sequence the human genome[2]
that ran alongside the Human Genome Project. He established the J. Craig Venter Institute in 2006, and in 2010 announced that he had succeeded in creating a synthetic cell, built from a chemically synthesised genome and capable of self-replication[3]
. As Farah puts it: “What Craig has been able to do is go from reading the genome to writing the genome.”
What Craig has been able to do is go from reading the genome to writing the genome
As the science has progressed, so has the technology underpinning it. A 60-kilobase viral genome can now be synthesised in a day; this was simply not possible a few years ago. “Megabase genomes in bacteria would have taken years to synthesise — now we can do this in weeks or months,” says Farah. This speed is accelerating the cycle of design, build and test that is so essential to synthetic biology. “It’s going to have a big impact on discovery and development,” he adds.
Synthetic Genomics says that it can now create vaccine seeds for virtually any strain of flu within a couple of days.
Drugs on demand

Source: Amyris
Jack Newman (pictured) is one of the founders of Amyris, a company that engineers yeast cells to produce pharmaceuticals, including a precursor of the anti-malarial drug artemisinin
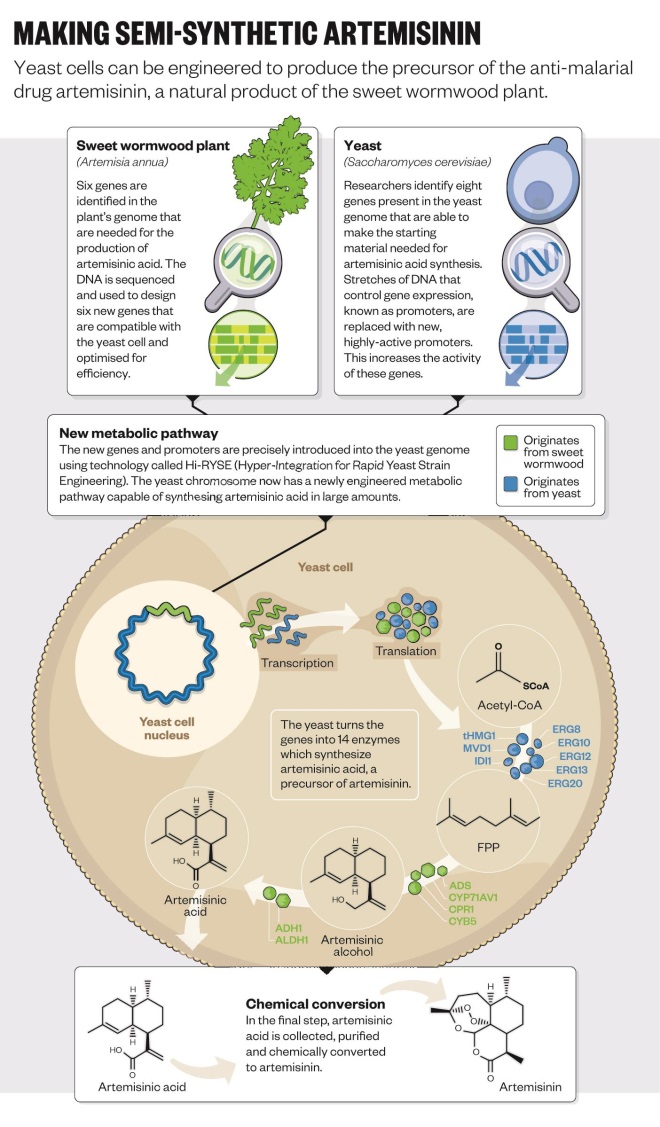
So far, one of the biggest successes of synthetic biology has been the antimalarial drug artemisinin. The drug is produced naturally by the Chinese sweet wormwood plant,
Artemisia annua, but only in tiny amounts. An engineered strain of yeast can now produce the drug on demand[4]
and is “infinitely better than the plant” at making it, says Jack Newman, co-founder and senior adviser at Amyris, a microbial engineering company based in Emeryville, California.
Amyris, together with the University of California, Berkley, developed the technology and retains the intellectual property rights to the synthetic biology platform. But pharmaceutical company Sanofi is now licensed to manufacture artemisinin for supply to the developing world on a non-profit basis. Before the new technology, artemisinin production was slow because it takes about a year to grow a crop of sweet wormwood. “If you were a contractor in Africa it used to take 18 months to actually get the drug after placing the order,” says Newman. There were constant shortages, which led to a cycle of over- then underproduction, he explains. “Now we have a stable source that can produce the world’s requirement in a month.” It is also much cheaper, he adds.
But developing the yeast-based platform was not straightforward (see ‘Semi-synthetic arteminisin’). It began at the University of California, Berkeley, in Jay Keasling’s laboratory, which has a long and distinguished history in synthetic biology. To take the technology forward, Amyris was founded in 2003 by four postdoctoral students at Berkley and Keasling. “We realised there was something happening in biology that could advance pharmaceutical developments,” says Newman, one of those original postdocs. “At the time we called it ‘metabolic toolchest’, but it wasn’t quite as catchy as synthetic biology.”
We realised there was something happening in biology that could advance pharmaceutical developments
Newman says that he and his colleagues would get together after work, eat Chinese food and pitch ideas about how they could use this new science for the greater good. “We saw that developing it to produce artemisinin could have a huge impact, but there wasn’t much interest because [malaria is] a problem of the developing world.” But in 2005 they received funding from the Bill & Melinda Gates Foundation.
At the time, no one had ever engineered an organism to produce an entirely foreign drug pathway of this complexity, and Newman reckons the odds of it working were 1,000 to 1. The first thing to do was find the genes in the sweet wormwood plant that encode the enzymes needed to synthesise artemisinin — but none of these genes had been identified yet. “UC Berkley had an army of postdoctoral students and PhDs working on this,” he says. After a year and a half, they had all the genes needed to make artemisinic acid in yeast, and they could convert this to artemisinin. But it was only when Patrick Covello, now a senior research officer at the National Research Council Canada’s Plant Biotechnology Institute, identified two more genes that production became quick enough to make the system commercially viable.
Optimisation of the system took two years. At which point Amyris joined forces with Sanofi to scale up the manufacturing and distribution. Now Amyris has the most advanced synthetic biology platform for producing pharmaceuticals, known as the ‘µPharm’
and uses it to make products including clean, green biofuels and the skin moisturiser squalene. A team at Stanford University has since successfully engineered yeast to produce the opioid drugs thebaine and hydrocodone, which could previously only be sourced from the poppy plant[5]
.
Re-engineering yeast to produce different chemicals is like writing a new app for an iPhone, says Newman. “So you might say, right, I want a molecule that looks like this, and then you go and find the genes that encode the enzymes for that chemical process.” Newman describes enzymes as tiny little machines that precisely join carbon to carbon. In contrast, he says that chemical synthesis of a molecule is like “mixing a bunch of wood and nails together and shaking them up and getting a house”.
Editorial advice: Amyris
Technology of the future

Source: Wyss Institute
Jeffrey Way at the Wyss Institute, Harvard, has just been given funding to engineer a network of bacteria that can be used to treat gastrointestinal infections
In synthetic biology, nothing is left to chance. “You get one shot,” says Jeffrey Way, who develops therapeutic proteins at Harvard’s Wyss Institute in Boston, Massachusetts. It is “the opposite of trial and error”, he explains. “Everything happens because we program it to.”
If drugs are engineered to behave more precisely, with two or three complementary functions, for example, then they could do more, says Way. But this is incompatible with the high-throughput techniques used in industry. “You need a fundamentally different approach,” he says. It requires the scientists to model each function of the drug and calculate how they interact.
Way builds drugs with two or three different components and has had some success with erythropoietin — a hormone that stimulates the bone marrow to make more red blood cells. The erythropoietin receptor also exists on other cells, which can cause unwanted effects if erythropoietin binds to these. So Way combined a mutated erythropoietin gene with the gene for a fragment of an antibody. The mutated hormone has less affinity for its receptor, which seems counterintuitive. However, “the antibody therefore determines what the drug binds to and targets the protein directly to the red-blood-cell precursors, limiting the side effects,” explains Way[6]
. This approach has subsequently been applied to interferon-alpha and other cytokines. Way runs thousands of iterations of the protein through a supercomputer that calculates how the drug will behave in the human body.
Way’s lab, which he runs with his wife, Pam Silver, has recently been awarded a grant by the Defense Advanced Research Projects Agency (DARPA), which is based in Arlington, Virginia. The lab has been tasked with developing bacteria that can be taken like a pill to provide protection from traveller’s diarrhoea — something that invariably afflicts soldiers when they are deployed abroad. The aim is to engineer the bacteria to sense the gastrointestinal inflammation caused by pathogenic bacteria and to treat it by killing them and releasing anti-inflammatory drugs. The team won the grant by showing that they could permanently switch on a gene in bacteria in the guts of mice by administering a drug[7]
.
Funding from DARPA is focused on advanced science and technology that has applications for the military. The agency’s previous successes include the development of both the internet and global positioning system (GPS) technology. But DARPA is increasingly turning its attention to biology, and recently set up the DARPA Biological Technologies Office. As Way puts it: “Biology is the technology of the future — carbon is the silicon of this century.”
Biology is the technology of the future — carbon is the silicon of this century
And DARPA is pushing hard to help scientists reach these new horizons. It will give the Wyss team up to US$4.7m for their work on traveller’s diarrhoea, for example. “What I like about DARPA is that they force you to be different and they also give you lots of money,” says Way. But at the same time they set a very high bar and expect people to fail. “If the research they fund doesn’t fail a lot of the time then they think they’re not taking big enough risks,” he says.
For the traveller’s diarrhoea project, DARPA has mandated that there must be five different species of bacteria that work together to sense and treat gut inflammation. “In this system, when the five species are together they can grow, but when they’re apart they will die. If there was just one species it may be able to evolve,” explains Way. So the team at Wyss must engineer a community of five co-dependent species of bacteria that also behave like a diagnostic and a drug — not an easy feat. “This was DARPA’s decision and everyone is challenged by it,” says Way.
But DARPA has good reason for imposing this complexity. There are ethical considerations when synthetic biology is used in this way. The concern here is that the mutant species created could wreak havoc in the wild. “DARPA wants organisms that are safe to release in the environment — they must die when they’re in the wrong place, and they must not exchange DNA with other organisms,” says Way. With some clever programming, he believes that he can stop them escaping into the wild, although he thinks that the threat to the environment is hypothetical.
Out of this world
Maintaining the health of the gut microbiome is not just essential for soldiers — it is also important for astronauts. “Understanding the microbiome is incredibly important to human health, so being able to manipulate it while astronauts are in space would be invaluable,” says John Hogan, an environmental scientist at NASA’s Ames Research Center in Moffett Field, California.
At the moment, NASA is leaving this research in the capable hands of institutions such as Wyss. But it does have its own aspirations in synthetic biology. “Mars has been a desired target for space travel for a long time, but we think it will be a three-year round trip, and keeping people healthy for this long is going to be difficult,” Hogan explains. The Ames Research Center has recently invested in a programme to produce ‘nutraceuticals’ for astronauts, for example to protect them from the higher levels of radiation in space.
“One of the biggest challenges for a mission to Mars is the number of different nutrients you need,” says Hogan. “How do you meet the astronaut’s requirements for three years? If we can’t bring them all with us then we have to make some of them.” Researchers at NASA are working on a yeast-based system. The yeast would be transported as spores, which can be stored for a long time and are resistant to environmental change. Hogan imagines the spores being kept in something like a takeaway ketchup packet. “You’d rehydrate them when needed,” he says.
Synthetic biology appeals to NASA because it offers a unique level of control over biological systems, and these systems are incredibly efficient and are able to make things that physical or chemical systems just cannot, says Hogan. “Every microorganism is a finely tuned chemical synthesis platform, and it replicates itself over and over again pretty reliably,” he says. “If we were trying to create a system like this from scratch, we’d be creating life!”
If we were trying to create a system like this from scratch, we’d be creating life!
But Hogan says that in some circumstances a biological system may not be the best option. “NASA has a strong interest in producing medicines on demand. This could be a chemical platform, it just depends on which is better suited,” he says. For this reason, NASA is keeping a keen eye on a DARPA-funded project called Battlefield Medicine. The aim of the project is “to develop miniaturised device platforms and techniques that can produce multiple small-molecule active pharmaceutical ingredients and therapeutic proteins in response to specific battlefield threats and medical needs as they arise”.
Grants for the project have gone towards the development of both chemical and biological systems. One of the centres that has received funding is the Massachusetts Institute of Technology (MIT), which is working on the biological system. Chris Love, a chemical engineer, who is leading the project at MIT, says that it addresses the broader question of “how to get medicines to patients when and where they need them”. For example, during a disaster, such as Hurricane Katrina, which hit Louisiana in 2005, thousands of patients needed insulin, but there were logistical problems, such as a lack of cold storage, and lots of insulin went to waste, says Love. So his team is trying to make a portable system that can produce biological drugs on demand within 24–48 hours.
“We’ve made a couple of molecules and shown feasibility,” says Love, who has just secured two more years of DARPA funding. Currently, many therapeutic proteins such as monoclonal antibodies are produced by mammalian cell systems, but Love’s team, like a number of others, has chosen a yeast-based system. “Microbial systems replicate very quickly, and as synthetic biology advances we can engineer yeast to produce more humanised proteins,” he says.
Microbial systems replicate very quickly, and as synthetic biology advances we can engineer yeast to produce more humanised proteins
One requirement is that the system must be small and easy to transport. “Originally the objective was to fit it into a backpack, but now it’s looking like it will be something you can put in the back of a car or a jeep,” Love says. He has high hopes for what such a small-scale, agile manufacturing unit could mean for medicine and research. For a start it would be significantly cheaper than large manufacturing units that cost hundreds of millions of dollars. And it means that small amounts of drugs, which might be needed to treat rare diseases, for example, would be much easier to produce and take to patients.
Love also thinks that the system could be applicable to early stage clinical trials. Imagine that a doctor working in a hospital thinks she has discovered a new therapeutic antibody and has promising data from a mouse study. Instead of having to take the finding to a pharmaceutical company, the doctor can identify the relevant genes and send the DNA sequence to a biotech company, which can send back a custom-made manufacturing system that produces her antibody on demand. And the system can be located on-site at the hospital. “Wouldn’t that be amazing?” asks Love.
Ultimately, however, the true power of synthetic biology may not be in medicine. “It will absolutely make a huge difference to medicine, but I almost see this as low-hanging fruit,” says Newman at Amyris. His company has successfully produced synthetic, renewable biofuels. But because biofuels are more expensive than petrol, it is hard for them to make an impact. “In 100 to 150 years, the way we source products will have to fundamentally change when the world’s oil runs out, otherwise there will be war and famine on a scale never seen before,” he says. “Synthetic biology will have a big impact, but it also has to.”
References
[1] Fleischmann RD, Adams MD, White O et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 1995;269:496–512. doi:10.1126/science.7542800
[2] Venter JC, Adams MD, Myers EW et al. The sequence of the human genome. Science 2001;291:1304–1351. doi:10.1126/science.1058040
[3] Gibson DG, Glass JI, Lartigue, C. et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 2010;329:52–56. doi:10.1126/science.1190719
[4] Paddon CJ, Westfall PJ, Pitera DJ et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013;496:528–532. doi:10.1038/nature12051
[5] Galanie S, Thodey K, Trenchard IJ et al. Complete biosynthesis of opioids in yeast. Science 2015;349:1095–1100. doi:10.1126/science.aac9373
[6] Taylor ND, Way J, Silver PA et al. Anti-glycophorin single-chain Fv fusion to low-affinity mutant erythropoietin improves red blood cell-lineage specificity. Protein Engineering, Design and Selection 2015;23:251–260. doi:10.1093/protein/gzp085
[7] Kotula JW, Kerns SJ, Shaket LA et al. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proceedings of the National Academy of Sciences USA 2014;111:4838–4843. doi:10.1073/pnas.1321321111