Human Protein Atlas
A map showing the locations of almost 17,000 different proteins in the human body could offer invaluable guidance for drug developers.
The Human Protein Atlas published the first major analysis of its data in Science
[1]
on 23 January 2015, including the whereabouts of the proteins targeted by every approved drug on the market.
The atlas is free to access via an interactive website
that allows researchers to quickly navigate the proteins found in 32 tissue types, representing all the body’s major tissues and organs.
“Researchers can come to find their favourite protein, and where it is located in the body,” says Tove Alm from the Science For Life Laboratory in Stockholm, who is a member of the Human Protein Atlas team. She adds that the site received 53,000 unique visitors when it launched in November 2014. “We’ve worked on this for about 11 years,” says Alm, “and everyone is very excited that we’ve got this far — it’s been a real team effort.”
Almost all pharmaceuticals work by controlling the activity of proteins. “So it’s extremely useful information for drug development, because we now know where our targets are expressed,” says Hanno Langen, global head of protein and metabolite technologies at F Hoffmann-La Roche in Basel, Switzerland. According to the DrugBank database, all of the drugs approved by the US Food and Drug Administration collectively target some 620 proteins, including enzymes, transporters, ion channels and receptors.
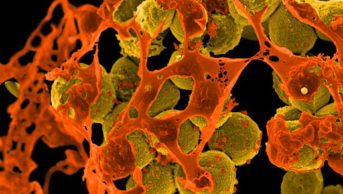
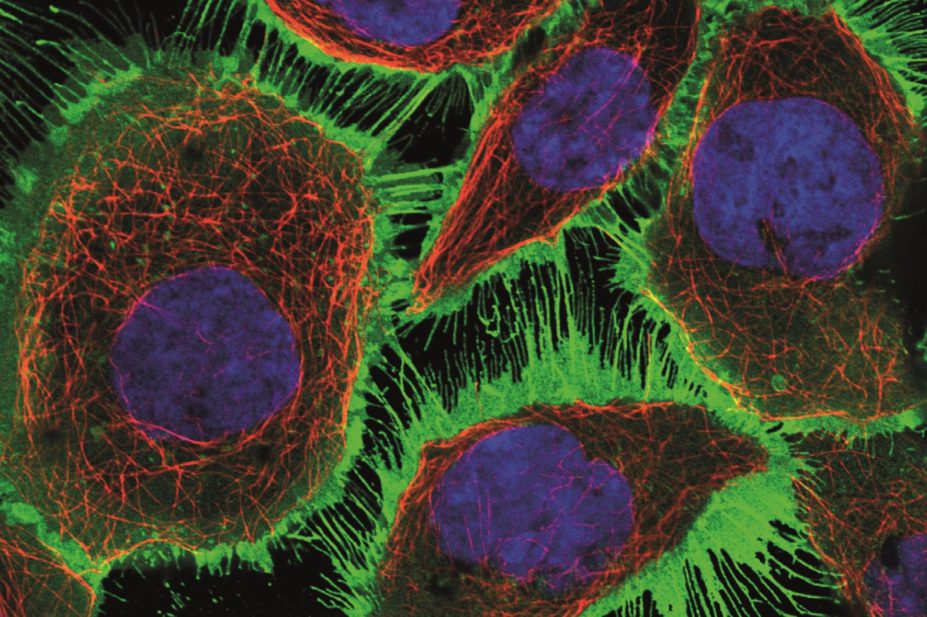
The Human Protein Atlas researchers created their database by working through a list of protein-coding genes in the body, and using the relevant DNA sequences to generate antibodies that would bind to their complementary proteins. They tested more than 24,000 of these antibodies in tissue samples and cell lines, imaging the bound proteins in cells using fluorescent tags and other techniques (see image). The team also sequenced the RNA in each of the 32 tissue types, to measure the activity of each protein-coding gene.
The distribution of the proteins is recorded in 13 million annotated images, showing their location down to the single-cell level — and in many cases, their distribution within individual cells. “I use it frequently,” says Gilbert Ommen of the University of Michigan, Ann Arbor, who studies the role of proteins in cancers. “It is the definitive source of information about protein expression.”
The atlas covers around 85% of the estimated 20,000 protein-producing genes in humans. The remaining genes might be expressed in tissues that have not yet been surveyed, including the eye or parts of the brain. “These are tissues we really hope to work on in the future,” says Alm.
According to the atlas, almost 9,000 protein-coding genes are expressed throughout the body, suggesting that they have core ‘housekeeping’ functions such as protein synthesis, energy generation or gene expression. Around 30% of the proteins that are targeted by drugs have this widespread distribution, which may explain some of the drugs’ systemic side effects.
Ommen says that researchers can now use the atlas to predict where these side effects are likely to arise, giving them a much better chance of identifying such problems earlier in the drug-discovery process.
The atlas also shows that roughly 15% of our protein-coding genes are expressed in a much more localised way, creating proteins that reside in specific tissues or organs. Focusing on these proteins might offer a way to develop better-targeted drugs, says Alm.
The team is planning further data releases in the coming years, including chapters of the atlas that will compare different areas of the brain, and map the sub-cellular locations of proteins in more detail.