IDbyDNA Inc.
High-throughput genetic sequencing can help to identify pathogens during diagnosis and treatment of infections. But applying the technology in practice has been held back by slow computation times and analytical inaccuracies.
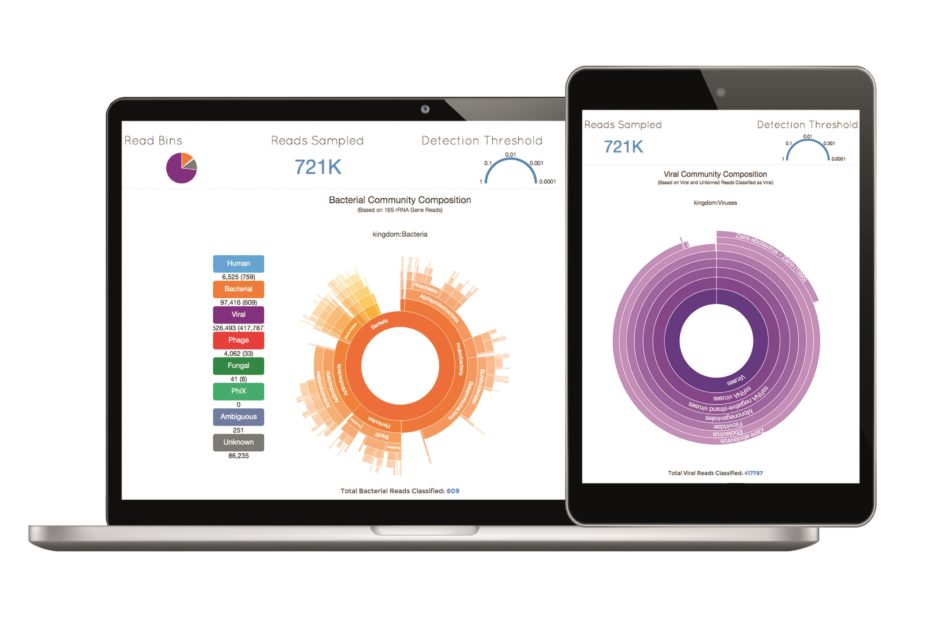
A team from the United States has developed a web tool called Taxonomer that can analyse metagenomic DNA and RNA data at speeds 10 to 100 times faster than currently available software. It can detect any known virus, bacterium or fungus, as well as discover novel pathogens.
Presenting their findings in Genome Biology (online, 26 May 2016)[1]
, the researchers say the tool was able to detect unrecognised pathogens in patients who had previously tested negative and correctly identify microbial contamination within stem cell cultures.
The researchers say that the free tool, which can be used on computers and mobile devices, will allow rapid data sharing in response to disease outbreaks.
References
[1] Flygare S, Simmon K, Miller C et al. Taxonomer: an interactive metagenomics analysis portal for universal pathogen detection and host mRNA expression profiling. Genome Biology 2016. doi: 10.1186/s13059-016-0969-1


