
Photoshot Holdings Ltd / Alamy
If you try to imagine a disease outbreak investigation you’ll probably think of something from a Hollywood blockbuster: a world-renowned virologist being helicoptered by the military as they race against time to find the person who is the unwitting source of the infection, and within them the blood sample that may lead to a cure.
But according to David Heymann, an infectious-disease epidemiologist at the London School of Hygiene & Tropical Medicine, it’s not quite like that. “Finding patient zero may be important in some instances,” he says, “but only if they are still alive and spreading the disease. And more often than not, especially in large disease outbreaks, they’re not.”
Classic epidemiological investigations typically involve laborious detective work, but advances in molecular techniques mean that epidemiologists no longer have to knock on quite so many doors to find their patient zero.
Contact tracing
An outbreak investigation begins when a patient presents to a hospital with an illness. Hospital staff record a history of the patient’s recent contacts to find out whether others are in need of medical treatment, but also to help diagnose the disease. Epidemiologists also need clues about the infection’s transmission course and how to stop it spreading further.
Looking at that patient and taking their history led to other contacts who had been unwell
“Take Kikwit in 1995,” Heymann says, referring to an Ebola outbreak in the city in the southwest of the Democratic Republic of the Congo that killed an estimated 245 people. Heymann was a member of the team that had investigated the first known outbreak of the disease in Yambuku in the north of the country in 1976, and he led the Kikwit investigation two decades later. “The first patient in the Kikwit outbreak was identified at the beginning of March. Looking at that patient and taking their history led to other contacts who had been unwell,” Heymann says. “We traced the outbreak back to having probably originated in January in a person who worked in the forest making charcoal.”
Heymann muses that once the forest worker was infected, probably by bats, there was a small, inconspicuous chain of transmission in February until the virus reached a hospital, where it was spread by poor practices such as needle sharing. “Health workers began to get infected and started to take the disease out into the community. From there the virus spread in a much more explosive manner than it ever would have done in just a person-to-person transmission chain.”
Once Heymann’s team realised that the outbreak was driven by poor hospital practices, they knew where to intervene. It also gave them a focal point to help them trace further patients and implement control strategies, including isolation and education. They managed to wrestle the outbreak to a halt.
The most infamous patient zero is Mary Mallon, better known by her unfortunate moniker of Typhoid Mary. Mallon was a cook in New York state and was an asymptomatic carrier of Salmonella typhi, the bacterium that causes typhoid fever. Between 1900 and 1915, while working for affluent families, hotels and hospitals, she is thought to have transmitted the pathogen to about 50 people, at least three of whom died.
Tracking the illness back to Mallon required arduous epidemiological investigation and legal intervention — twice — to stop her from spreading the disease. But epidemiologists can now use molecular techniques to track patient zero more quickly.
Sequencing technology
Since the advent of high-throughput genomic sequencing in 2005, techniques that once required six-figure budgets and years of manpower have become available to ordinary epidemiologists.
“In the past few years, a number of companies have developed sequencing platforms about the size of a large cappuccino machine that sit on the laboratory bench,” says Estée Török, a microbiologist at the University of Cambridge and a consultant in infectious diseases and microbiology at Addenbrooke’s Hospital, Cambridge, UK. “These machines can sequence a bacterial genome in less than a day and at a very reasonable cost — around £70 per genome. This means that we are able to use the technology in real time to investigate outbreaks.”
Once you are able to identify the source, everyone wants to know who it is and they start pointing fingers
In 2011, Török was confronted with such an opportunity. Over a period of six months, 12 babies in the neonatal intensive care unit of Addenbrooke’s Hospital were infected with methicillin-resistant Staphylococcus aureus (MRSA). Normal investigational techniques using epidemiological data and antibiotic susceptibility testing did not initially detect the outbreak, but Török and her colleagues used rapid bacterial whole-genome sequencing and were able to confirm the presence of a persistent outbreak. They were able to identify transmission to other babies, mothers and relatives on other wards and in the community. They traced the infection back to a health worker who was found to be carrying the outbreak strain. This individual was treated and the outbreak was brought to a close[1]
.
“Once you are able to identify the source, everyone wants to know who it is and they start pointing fingers, which is unfortunate,” says Török. She refers to the 2010 outbreak of cholera in Haiti in which similar sequencing techniques identified the source as United Nations (UN) troops from Nepal, a finding that led to riots outside the UN base. “From our point of view, it doesn’t matter who patient zero was,” Török says. “The good news is that we can use this technology in real time to identify the source of infection, intervene and terminate the outbreak before it can spread further. These are very exciting times in the field.”
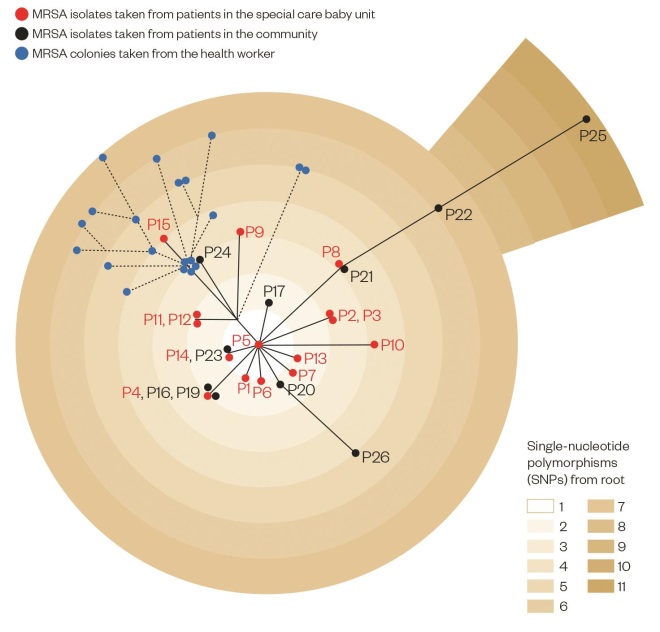
Mapping the Addenbrooke’s Hospital MRSA outbreak
Source: Source: Harris SR, Cartwright E, Török ME et al. Lancet Infect Dis;2013:130–136
In this phylogenetic analysis, genome sequencing was used to map the connections between samples taken from babies in the special care baby unit (Patients (P)1–15, shown in red) and from people in the community (P16–P26, black). 20 individual MRSA colonies (blue) were taken from a health worker towards the end of the outbreak. Each gradation in colour shows one single-nucleotide polymorphism, or mutation, and thus genetic distance from the reference MRSA sequence type 22 strain.
Global intelligence
The excitement stems from the rapidly decreasing cost and increasing availability and sophistication of genomic sequencers. Over the past decade, sequencing capacity has doubled every six to nine months. Since 2007, the cost of sequencing an entire genome has plummeted from around US$10m (£7m) to about US$7,000 in 2014, and the cost per million base pairs of sequence has fallen from nearly US$1,000 to less than 10 cents[2]
.
The molecular approach to studying specific agents associated with disease has brought another tool to help improve the effectiveness of the classic epidemiological investigation
Putting more sensors in the hands of front-line doctors means there is a better chance of detecting patient zero while he or she is still relevant to an outbreak, and it also allows for more sensitive and intelligent global surveillance systems.
“The molecular approach to studying specific agents associated with disease has brought another tool to help improve the effectiveness of the classic epidemiological investigation,” says Mark Pallansch, director of the Division of Viral Diseases at the Centers for Disease Control and Prevention in Atlanta, Georgia. “We don’t use either in isolation anymore. It’s routine to have a multidisciplinary approach to investigating an outbreak.”
The molecular approach has widened the possibilities for what epidemiological investigations can achieve. “In some situations, like in polio with its many undetected infections for every case, without molecular epidemiology it would not be possible to trace back patient histories,” Pallansch says, pointing to the response to the 2013 detection of polio in Syria. In that outbreak, sequencing data collected by the Global Polio Laboratory Network identified the outbreak as a strain that originated in Pakistan, so containment and vaccination operations could be localised around its likely route.
Molecular epidemiology provides predictive capabilities too. Data collected by national surveillance laboratories feed into overarching surveillance networks, such as the World Health Organization’s (WHO) Global Influenza Surveillance and Response System (GISRS), information from which determines the make-up of seasonal influenza vaccines. Other networks, such as the Global Public Health Intelligence Network (GPHIN), along with smaller, web-based, report-aggregating systems such as ProMed-Mail and Health Map, can provide the first warnings of disease outbreaks for national authorities and then arm public-health practitioners with the epidemiological information they need to tackle the outbreak.
It was ProMed-Mail and GPHIN that first alerted health authorities to an outbreak of a highly fatal respiratory disease that originated in Guangdong Province in China in 2002 and was later named severe acute respiratory syndrome (SARS). Investigators tracked the SARS pandemic back to a medical doctor who had travelled from Guangdong Province to Hong Kong and who became the index case there. He stayed at a hotel and infected the other guests, who then took the virus back to their home countries in Asia, Europe and North America, beginning the 2002–2003 SARS outbreak that killed nearly 800 people.
The SARS virus was identified when the GISRS could not find the influenza virus in specimens from a patient who had travelled from Hong Kong to Vietnam, ruling out influenza A subtype H5N1 or avian flu.
Heymann was assistant director-general at WHO at the time and was in charge of the SARS response. Armed with the knowledge of how the disease had spread, WHO and others acted to limit international travel and contained the outbreak before it could lay down roots in the human population. He says that the international community was lucky that the outbreak occurred in a developed region with a good health system that allowed such prompt detection and action.
Lessons from Ebola
That’s not been the case with the 2014 Ebola outbreak in West Africa, where health systems are fragile at best. The outbreak, which has so far killed more than 8,000 people, is thought to have originated in Meliandou, a remote forest village in southern Guinea. Its patient zero was a toddler who contracted the disease by unknown means and died from his illness in December 2013[3]
.
Given the uncertainty surrounding the epidemiology of the disease, says Donald Henderson, an epidemiologist from the Center for Health Security at the University of Pittsburgh, Pennsylvania, it is difficult to be sure that this child was indeed the source of the outbreak. “Another child or person with vomiting, fever and diarrhoea could have gone misdiagnosed,” he says, “at a time when nobody was particularly aware of, or concerned about, Ebola.”
He is sure, though, that there was a serious failure in reporting. WHO was notified of a mysterious disease, later identified as Ebola, in March 2014. By then the virus had found its way to the major trading centre of Gueckedou in Guinea, from where it spread into neighbouring Sierra Leone and Liberia, killing about 60 people. By late May 2014, with the outbreak unchecked, the virus had spread into major cities with millions of inhabitants, at which point it spiralled out of control.
“Why was it not detected, and why did it take so long for WHO to be informed?” asks Henderson. “I can’t answer that and I’m not sure anyone can.” He points to a separate Ebola outbreak in a rural region of the Democratic Republic of the Congo in August 2014. That outbreak, thought to have originated in a woman returning from an animal market, was identified and dealt with rapidly by health authorities — it was stopped in a matter of months with fewer than 70 lives lost.
If you have global surveillance networks, there is a safety net to tell you that an organism has not been dealt with properly and to allow the necessary control measures to take place
There are lessons to be learned from the recent Ebola outbreak. “At the global level we need to be working with each national government bilaterally to help strengthen their capacities,” says Heymann. He advocates channelling development assistance through the International Health Regulations, an international treaty that requires all countries to strengthen core public-health capacities, such as preparedness, surveillance, laboratory and response mechanisms, and risk communication, to help avoid international outbreaks. “Then, if you have global surveillance networks, which we have, there is a safety net to tell you that an organism has not been dealt with properly and to allow the necessary control measures to take place.”
Improving global capacity requires more than outside help, however. It also means developing capacity from within, says Pallansch. “Look at Ebola and the various influenza epidemics that have stemmed from rural locations. These things start relatively small at a local level before expanding and becoming major international concerns.” It would be foolish not to ensure that eventually every country has the capacity to detect potential outbreaks as soon as they occur, he says, adding: “In the meantime, if all countries do cooperate, I think we’re able to respond when and where needed.”
References
[1] Harris SR, Cartwright EJP, Török ME et al. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infectious Diseases 2013;13:130–136.
[2] National Human Genome Research Institute. Data from the NHGRI Genome Sequencing Program.
[3] Baize S, Pannetier D, Oestereich L et al. Emergence of Zaire Ebola Virus Disease in Guinea. New England Journal of Medicine 2014;371:1418–1425.


